This notebook contains code that exhaustively enumerates all possible conformations of an HP lattice protein. There are also functions to
count the number of topological contacts
compute the energy of a given conformation and sequence
select the compact conformations out of a set of generic conformations
given a sequence and a set of conformations, find the lowest-energy structures
use
matplotlibto show a conformation
At the end you can also find some example code that uses these functions.
import sys
import numpy as np
import random
import matplotlib.pyplot as plt
import itertools
import numpy as np# On a square lattice there are four possible directions (up, right, down, left)
DIRECTIONS = [(0, 1), (1, 0), (0, -1), (-1, 0)]
# Return all possible conformations of a given length
def generate_conformations(length):
# there is a single conformation of length 2 that is not related to others by rotations or translations
if length == 2:
return [[(0, 0), (0, 1)]] # this return stops the recursion of the next line
# Recursive call: get all possible conformations for length - 1
smaller_conformations = generate_conformations(length - 1)
conformations = []
for conf in smaller_conformations:
last_point = conf[-1]
for direction in DIRECTIONS:
new_point = (last_point[0] + direction[0], last_point[1] + direction[1])
if new_point not in conf: # check that there are no overlaps
# generate the new configuration
new_conf = conf + [new_point]
# generate its mirror image
mirror_conf = [(-x, y) for x, y in new_conf]
# pick one of the two using the lexographic order through min
conformations.append(min(new_conf, mirror_conf))
# make sure that only one of the two mirror images of each configuration is retained
conformations.sort()
return list(conformations for conformations, _ in itertools.groupby(conformations))
# Return the number of topological contacts in a conformation
def count_topological_contacts(conformation):
count = 0
length = len(conformation)
for i in range(length):
for j in range(i + 2, length): # + 2 to skip connected neighbours
if is_adjacent(conformation[i], conformation[j]):
count += 1
return count
# Return the conformations with the maximum compactness (i.e. with the maximum number of topological contacts)
def select_compact_conformations(conformations):
max_topological_contacts = 0
compact_conformations = []
for conf in conformations:
topological_contacts = count_topological_contacts(conf)
if topological_contacts > max_topological_contacts:
max_topological_contacts = topological_contacts
compact_conformations = [conf]
elif topological_contacts == max_topological_contacts:
compact_conformations.append(conf)
return max_topological_contacts, compact_conformations
# Return the energy of a conformation for a given HP sequence
def calculate_energy(conformation, sequence):
energy = 0
length = len(conformation)
for i in range(length):
if sequence[i] == 'H':
for j in range(i + 2, length): # + 2 to skip connected neighbours
if sequence[j] == 'H' and is_adjacent(conformation[i], conformation[j]):
energy -= 1
return energy
# Check if two points are adjacent in the lattice
def is_adjacent(point1, point2):
return abs(point1[0] - point2[0]) + abs(point1[1] - point2[1]) == 1
# Return the lowest energy conformations
def find_lowest_energy_conformations(sequence, conformations):
length = len(sequence)
if length != len(conformations[0]):
print(f"The length of the sequence ({length}) is different from that of the conformations ({len(conformations[0])})")
lowest_energy = float('inf')
lowest_energy_conformations = []
for conf in conformations:
energy = calculate_energy(conf, sequence)
if energy < lowest_energy:
lowest_energy = energy
lowest_energy_conformations = [conf]
elif energy == lowest_energy:
lowest_energy_conformations.append(conf)
return lowest_energy, lowest_energy_conformations
# Plot the conformation, with each residue coloured according to the sequence
def plot_conformation(conformation, sequence):
# define colors for H and P residues
color_map = {'H': 'blue', 'P': 'red'}
# used to shift the 'H' and 'P' labels so that they do not overlap with the symbols
label_shift = 0.05
# extract x and y coordinates
x_coords, y_coords = zip(*conformation)
# plot the conformation as a line connecting points
plt.plot(x_coords, y_coords, color='black', linestyle='-', marker='o', linewidth=2)
# plot each residue with its corresponding color
for i, (x, y) in enumerate(conformation):
plt.plot(x, y, marker='o', color=color_map[sequence[i]], markersize=10)
# the first residue is labelled differently to highlight where the chain begins
if i == 0:
label = sequence[i] + " (N)"
else:
label = sequence[i]
plt.text(x - label_shift, y + label_shift, label, fontsize=12, ha='right', va='bottom')
plt.axis('equal')
plt.grid(True)
plt.title(f"Conformation for sequence: {sequence}")
plt.axis('off')
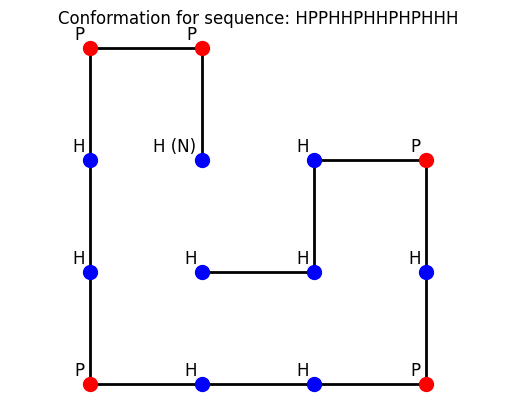
plt.show()sequence = "HPPHHPHHPHPHHH"
conformations = generate_conformations(len(sequence))
max_topological_contacts, compact_conformations = select_compact_conformations(conformations)
lowest_energy, lowest_energy_conformations = find_lowest_energy_conformations(sequence, compact_conformations)
print(f"Number of total conformations: {len(conformations)}")
print(f"Number of compact conformations: {len(compact_conformations)} ({max_topological_contacts} topological contacts)")
print(f"Number of lowest-energy conformations: {len(lowest_energy_conformations)}")
print("Lowest Energy:", lowest_energy)
plot_conformation(lowest_energy_conformations[0], sequence)Number of total conformations: 110188
Number of compact conformations: 360 (7 topological contacts)
Number of lowest-energy conformations: 1
Lowest Energy: -7
def generate_all_sequences(length):
return [''.join(seq) for seq in itertools.product('HP', repeat=length)]
N = 10
conformations = generate_conformations(N)
max_topological_contacts, compact_conformations = select_compact_conformations(conformations)
print(f"Number of total conformations: {len(conformations)}")
print(f"Number of compact conformations: {len(compact_conformations)} ({max_topological_contacts} topological contacts)")
N_natives = []
for sequence in generate_all_sequences(N):
lowest_energy, lowest_energy_conformations = find_lowest_energy_conformations(sequence, compact_conformations)
N_natives.append(len(lowest_energy_conformations))
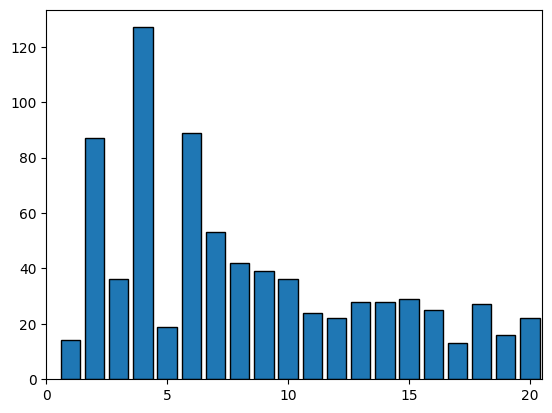
plt.xlim(0, 20.5)
plt.xticks(np.arange(0, 25, 5)) # set the tick label to multiples of 5
plt.hist(N_natives, bins=range(1, max(N_natives) + 1), ec='black', align='left', rwidth=0.8);Number of total conformations: 2034
Number of compact conformations: 98 (4 topological contacts)