In this notebook we study the dynamics of a particle evolving in a symmetric double-well potential to introduce the concepts of rare-event dynamics and transition-state theory.
The double-well potential has the form:
where we choose the constant so that both minima are exactly at zero energy. The barrier height is then .
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.animation import FuncAnimation
from IPython.display import HTMLHere we set the parameters and define the potential and its derivative
# Double-well potential parameters
a = 1.0
b = 4.0
V_shift = b*b/(4*a)
def V(x):
return a*x**4 - b*x**2 + V_shift
def dVdx(x):
return 4*a*x**3 - 2*b*x
m = 1.0 # mass
kT = 0.5 # thermal energy
nu = 1.0 # Andersen collision frequency (controls coupling strength)
dt = 0.002 # time step
steps = 300_000 # simulation length
sigma_v = np.sqrt(kT/m)Here we define a function that initialises the trajectory and one that generates the trajectory by using the Velocity Verlet integration algorithm, coupled to a simple Andersen thermostat
def init():
x = np.zeros(steps)
v = np.zeros(steps)
x[0] = -2.0 # start in left well
v[0] = 0.0
return x, v
def integrate(x, v, V_der, steps):
rng = np.random.default_rng()
for i in range(steps - 1):
# --- Velocity Verlet ---
# first step
v_half = v[i] - 0.5 * dt * V_der(x[i]) / m
# position integration
x[i+1] = x[i] + dt * v_half
# second step
v_new = v_half - 0.5 * dt * V_der(x[i+1]) / m
# --- Andersen thermostat randomization ---
if rng.random() < nu * dt:
v_new = rng.normal(0, sigma_v)
v[i+1] = v_newNow we run the simulation
x, v = init()
integrate(x, v, dVdx, steps)Now we plot the energy as a function of time
plt.figure(figsize=(12,4))
plt.plot(np.arange(steps)*dt, x, lw=0.7)
plt.axhline(0.0, color='red', ls='--', lw=1.2, label='Barrier (TS region)')
plt.ylim(-3, 3)
plt.xlabel("Time")
plt.ylabel("Position x(t)")
plt.title("Langevin Dynamics in a Double-Well Potential")
plt.legend()
plt.show()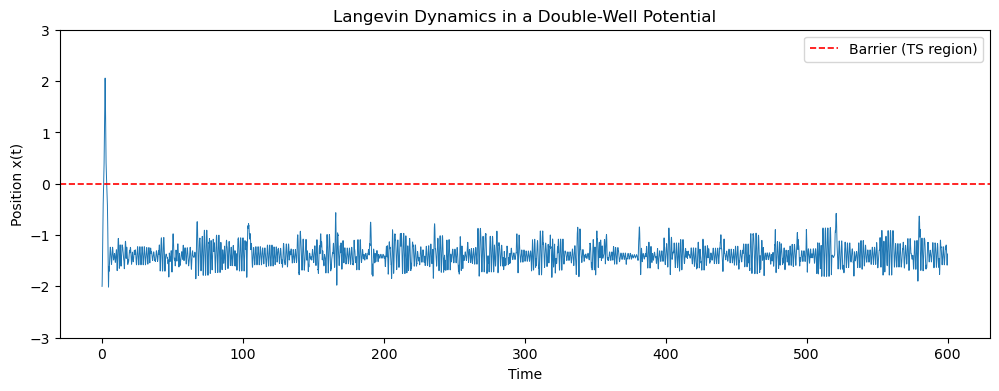
And then prepare an animation showing the position of the particle as a function of time using the trajectory we just generated. Here we first define a function so that we can use it later
def get_animation(xmin, xmax, V_pot, x):
# Create grid for potential
xmin = -2.5
xmax = 2.5
xs = np.linspace(xmin, xmax, 400)
betaVs = V_pot(xs) / kT
fig, ax = plt.subplots(figsize=(6,4))
ax.plot(xs, betaVs, 'k', lw=2)
particle, = ax.plot([], [], 'ro', ms=8)
ax.set_xlim(xmin, xmax)
ax.set_ylim(min(betaVs)-1, max(betaVs)+1)
ax.set_xlabel('x')
ax.set_ylabel(r'$\beta$ V(x)')
ax.set_title('Particle in Double-Well Potential')
def init_animation():
particle.set_data([], [])
return particle,
def update(frame):
particle.set_data(x[frame], V_pot(x[frame]) / kT)
return particle,
anim = FuncAnimation(fig, update, frames=np.arange(0, steps, 1000),
init_func=init_animation, blit=True)
plt.close()
return animShow the animation relative to the “regular” simulation
anim = get_animation(-2.5, 2.5, V, x)
HTML(anim.to_jshtml())/tmp/ipykernel_3542950/318829961.py:22: MatplotlibDeprecationWarning: Setting data with a non sequence type is deprecated since 3.7 and will be remove two minor releases later
particle.set_data(x[frame], V_pot(x[frame]) / kT)
1Umbrella Sampling Around the Barrier Top¶
In unbiased simulations, configurations near the top of the barrier are visited very rarely, making it hard to estimate the free-energy profile there.
To improve sampling of these rare configurations, we use Umbrella Sampling (US):
We add a harmonic bias centered at the barrier top :
This stabilizes the particle near the barrier region.
From the biased simulation, we collect a histogram .
We then unbias the distribution:
and reconstruct the free energy
In 1D, the theoretical free energy (up to a constant) is simply
so we can directly compare the US reconstruction to the known potential.
We start by defining some parameters, as well as the biasing functions
# --- Umbrella (harmonic) bias centered at the barrier top x0=0 ---
k_bias = 10.0
x0 = 0.0
def V_bias(x):
return 0.5 * k_bias * (x - x0)**2
def dVbias_dx(x):
return k_bias * (x - x0)
def V_total(x):
"""Total potential used in the umbrella simulation."""
return V(x) + V_bias(x)
def dVtotal_dx(x):
"""Derivative of the total potential used in dynamics."""
return dVdx(x) + dVbias_dx(x)And then run the simulation with the functions defined before
x, v = init()
integrate(x, v, dVtotal_dx, steps)Generate the animation
anim = get_animation(-2.5, 2.5, V, x)
HTML(anim.to_jshtml())/tmp/ipykernel_3542950/318829961.py:22: MatplotlibDeprecationWarning: Setting data with a non sequence type is deprecated since 3.7 and will be remove two minor releases later
particle.set_data(x[frame], V_pot(x[frame]) / kT)
Build biased histogram and unbias it
# Optional (for such a simple system): discard initial part (equilibration)
burn = steps // 5
x_samples = x[burn:]
# Histogram range and bins
nbins = 80
xmin, xmax = -2.5, 2.5
hist, bin_edges = np.histogram(
x_samples,
bins=nbins,
range=(xmin, xmax),
density=True # -> hist approximates pdf P_bias(x)
)
bin_centers = 0.5 * (bin_edges[:-1] + bin_edges[1:])
# Biased pdf P_bias(x)
P_bias = hist
# Bias potential at bin centers
Vb_centers = V_bias(bin_centers)
# Unbias:
# P_unbiased(x) ∝ P_bias(x) * exp(+beta * V_bias(x))
# Free energy: F(x) = -kT ln P_unbiased(x) + const
# Note: F(x) = -kT ln P_bias(x) - V_bias(x) + const
F_biased = np.full_like(P_bias, np.nan, dtype=float)
F_unbiased = np.full_like(P_bias, np.nan, dtype=float)
mask = P_bias > 0
F_biased[mask] = -kT * np.log(P_bias[mask])
F_unbiased[mask] = -kT * np.log(P_bias[mask]) - Vb_centers[mask]
# Theoretical free energy (up to constant): F_th(x) = V(x)
xs = np.linspace(xmin, xmax, 400)
# recalling that we can always shift a free energy arbitrarily,
# we shift the three free energy profiles so that the can be more
# easily plotted on the same figure
# Set the minimum of the unbiased free energy to zero
F_biased -= np.nanmin(F_biased)
# Set the maximum of the unbiased free energy to zero
F_unbiased -= np.nanmax(F_unbiased)
# Shift the maximum of the theoretical free energy to zero
F_th = V(xs) - V_shiftplt.figure(figsize=(7,4))
# Theoretical free energy
plt.plot(xs, F_th, label=r"Theoretical $F(x) \propto V(x)$", lw=2)
# Umbrella reconstruction (only where we have data)
plt.plot(bin_centers, F_biased, 'o', ms=4, label="US - biased")
plt.plot(bin_centers, F_unbiased, 'o', ms=4, label="US - unbiased")
plt.xlabel("x")
plt.ylabel("Free energy (arb. units)")
plt.title("Free energy near the barrier: umbrella vs theoretical")
plt.legend()
plt.grid(True, alpha=0.3)
plt.show()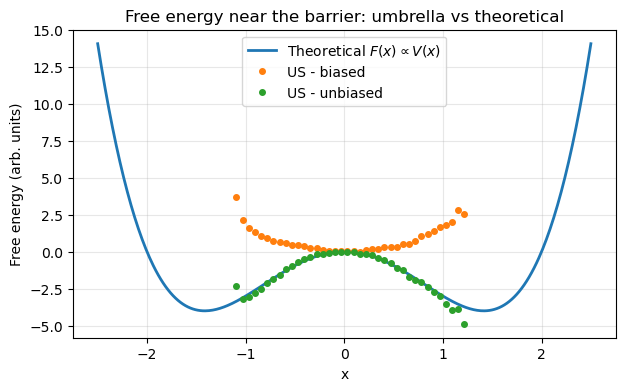
2Metadynamics: Turning Rare Events into Diffusion¶
Metadynamics is an enhanced-sampling technique designed to overcome free-energy barriers by building a history-dependent bias potential.
At regular intervals, small repulsive Gaussians are deposited in the space of a chosen collective variable (CV).
This discourages the system from revisiting already explored regions and gradually fills free-energy wells.
In this 1D example:
The collective variable is simply
Gaussians are deposited at the current particle position
The total potential becomes:
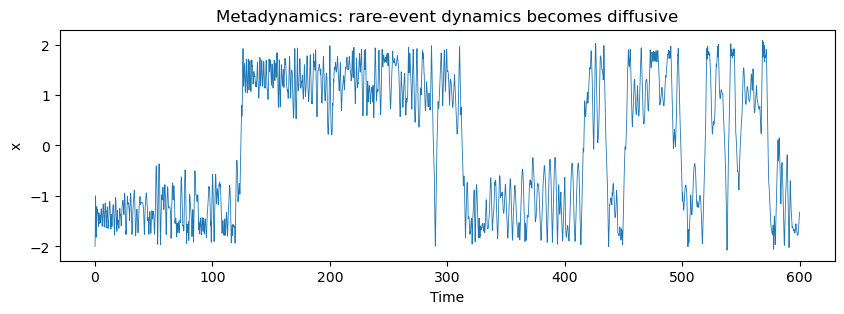
As time progresses, barrier crossings become frequent and the particle explores the full landscape.
For simple systems and long times:
# --- Metadynamics parameters ---
w = 0.05 # Gaussian height
sigma = 0.15 # Gaussian width
deposit_stride = 500 # deposit every N steps
# Grid for evaluating the metadynamics bias
x_grid = np.linspace(-3, 3, 400)
dx_grid = x_grid[1] - x_grid[0]
Vmeta = np.zeros_like(x_grid)def Vmeta_and_force(x):
"""
Evaluate metadynamics bias potential and force
using linear interpolation on a grid.
"""
# Keep x inside the grid
if x <= x_grid[0]:
return Vmeta[0], 0.0
if x >= x_grid[-1]:
return Vmeta[-1], 0.0
# Find grid index to the left of x
i = int((x - x_grid[0]) / dx_grid)
# Linear interpolation weight
t = (x - x_grid[i]) / dx_grid
# Bias potential: linear interpolation
Vb = (1 - t) * Vmeta[i] + t * Vmeta[i + 1]
# Force = - dV/dx = slope between grid points
Fb = -(Vmeta[i + 1] - Vmeta[i]) / dx_grid
return Vb, Fb
# --- Metadynamics simulation ---
x = np.zeros(steps)
v = np.zeros(steps)
x[0] = -2.0
v[0] = 0.0
rng = np.random.default_rng()
for i in range(steps - 1):
# --- Compute forces ---
# Physical force
F_phys = -dVdx(x[i])
# Metadynamics bias force
Vb, F_bias = Vmeta_and_force(x[i])
F_tot = F_phys + F_bias
# Velocity Verlet
v_half = v[i] + 0.5 * dt * F_tot / m
x[i+1] = x[i] + dt * v_half
# Recompute forces at new position
F_phys_new = -dVdx(x[i+1])
_, F_bias_new = Vmeta_and_force(x[i+1])
F_tot_new = F_phys_new + F_bias_new
v_new = v_half + 0.5 * dt * F_tot_new / m
# Andersen thermostat
if rng.random() < nu * dt:
v_new = rng.normal(0.0, sigma_v)
v[i+1] = v_new
# --- Deposit Gaussian bias ---
if i % deposit_stride == 0:
Vmeta += w * np.exp(-0.5 * ((x_grid - x[i]) / sigma)**2)
plt.figure(figsize=(10,3))
plt.plot(np.arange(steps)*dt, x, lw=0.6)
plt.xlabel("Time")
plt.ylabel("x")
plt.title("Metadynamics: rare-event dynamics becomes diffusive")
plt.show()
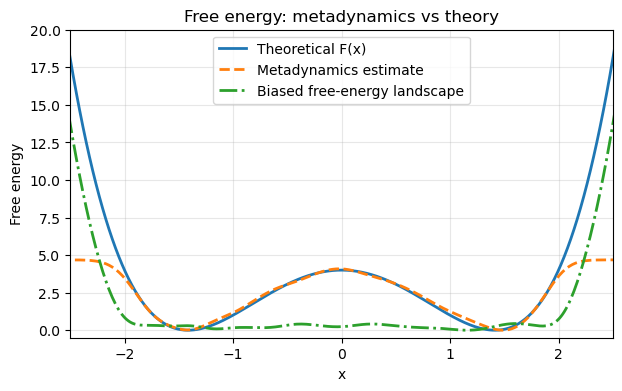
# Metadynamics free energy estimate
F_meta = -Vmeta
F_meta -= F_meta.min()
# Theoretical free energy
F_th = V(x_grid)
F_th -= F_th.min()
# Biased free energy
F_biased = F_th - F_meta
F_biased -= F_biased.min()
plt.figure(figsize=(7,4))
plt.xlim(xmin, xmax)
plt.ylim(-0.5, 20)
plt.plot(x_grid, F_th, label="Theoretical F(x)", lw=2)
plt.plot(x_grid, F_meta, '--', label="Metadynamics estimate", lw=2)
plt.plot(x_grid, F_biased, '-.', label="Biased free-energy landscape", lw=2)
plt.xlabel("x")
plt.ylabel("Free energy")
plt.legend()
plt.title("Free energy: metadynamics vs theory")
plt.grid(alpha=0.3)
plt.show()